Genes2WordCloud is a web-based server application and Java Applet that enables users to create biologically-relevant-content WordClouds.
A WordCloud is a visual display of a set of words where the font, size, color or angle can represent some underlying information. A WordCloud is an effective way to visually summarize information about a specific topic of interest. The WordCloud is optimized to maximize the display of the most important terms about a specific topic in the minimum amount of space.

As researchers are faced with the daunting amount of new and growing data and text, methods to quickly summarize knowledge about a specific topic from large bodies of text or data are critical. WordClouds are emerging as a method of choice on the web to accomplish this task.
Genes2WordCloud generates WordClouds from the following sources:
There are two tasks for creating WordClouds: first, generating the keywords to display; and secondly, displaying the keywords.
The keywords are generated in several ways depending on the source chosen. In each case the process can be divided into two main tasks: obtain the text related to the user input, and text-mine the text.

Diagram 1 - Main task 1: obtain text from the user input
Diagram 2 - Main task 2: text-mining

Diagram 3 -Text-mining task details
The Porter stemming algorithm is a common stemming algorithm which works well for English. It reduces words such as "stem", "stems", "stemming" to a single root, e.g., "stem". The root is not always a real English word. Therefore, to obtain a more readable WordCloud, after the stemming of all the words, each stemmed-word is replaced by the shortest word of its family.
It should be noted that some words are removed from the text before finding the keywords. First, all common English words such as: the, is, or are are removed. The list of these words can be found in the following file. Then common biological terms such as: experiments, abstracts, contributes are removed. These terms are available here. These terms were chosen by hand curation after experimenting with many WordClouds. Text-mining of generifs and gene ontology annotations also contains removed common terms. Finally, other terms such as the input gene names, the name of the author, or the keywords of the Pubmed search are also removed to avoid self-referencing.
The source files used to create the database for processing lists of genes to create WordClouds were taken from:
The different methods to obtain text from the user input and the text-mining algorithms consume a lot of CPU time and memory. For each query we only use a maximum of 150 abstracts or 500 annotations picked randomly when the queries return more than these limits.
There are currently two main web-based applications to create WordClouds from weighted lists of keywords: Wordle, developed by Jonathan Feinberg and indirectly IBM, and WordCram developed by Dan Bernier. Wordle cannot be used outside of the web application since its source code is protected, whereas WordCram is an open-source Java library using the Java libraries of Processing. Processing is a scripting language that uses Java Applets for creating web-based applications enriched with graphics. Genes2WordCloud is a WordCould viewer that is based on WordCram.
A web-based user-interface was added to Genes2WordCloud where several parameters such as the font or the background-color can be changed.
In this section we provide some examples of using Genes2WordCloud.
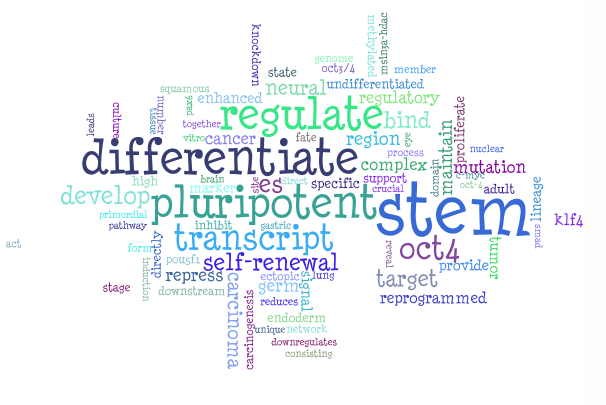
NANOG and SOX2 are both genes encoding transcription factors involved in embryonic stem cells self-renewal and pluripotency maintenance. The WordCloud automatically obtained relevant terms such as stem (the word cell was automatically removed as it is considered a biological common term), differentiate, pluripotent, self-renewal . Also Oct4, a gene that is often associated with NANOG and SOX2 was recovered by Genes2WordCloud.

The Ma'ayan Laboratory is a computational systems biology laboratory and the program correctly extracted the most relevant terms that describe the function of the lab, for example: network, mammalian, software, database, compute, web-based tool.

This WordCloud was obtained with the PubMed search: p38 pathway. The algorithm recovered terms such as: kinase, signal, MAPK, phosphorylate, apoptosis which are relevant to the p38 pathway, a signaling pathway involved in cell differentiation and apoptosis.
There are three possible explanations:
You can use the applet with your own keywords on your own website. For doing this all you need to do is:
These terms are stored in our database. If we agree that these should be indeed removed, we will add them to the common English words list or the common biological terms list.
Genes2WordCloud was developed by the Ma'ayan Laboratory , at Mount Sinai School of Medicine as part of the activities of the Systems Biology Center New York (SBCNY) .
If you have any particular issues, questions, remarks or suggestions, please Contact us.