
Hepatic fibrosis is a scarring response to liver damage often due to chronic liver disease. In fibrogenesis, hepatic stellate cells (HSCs) activate and transdifferentiate into ECM-producing myofibroblasts. To better understand the gene expression changes that occur during such a process, De Minicis et al.[1] conducted a microarray study to examine differences in gene expression profiles between cultured and in vivo activated HSCs. Using the study as a source of microarray data for X2K, we attempted to identify the upstream transcription factors, intermediate proteins, and protein kinases that may potentially regulate the fibrosis response in HSCs.
The differentially expressed genes found from the microarray study were split into two groups of up-regulated and down-regulated genes and sorted based on their fold change. We set the cut-off for up-regulated genes to have at least a fold change of 2 or greater, and for down-regulated genes to have at least a fold change of 0.7 or lower. The resulting lists for up-regulated genes and down-regulated genes has 469 genes and 660 genes respectively.
In the first stage of X2K analysis, the lists of up-regulated and down-regulated genes were each used to generate a list of predicted transcription factors. The transcription factor Tcf3 was predicted as top candidate that regulate changes in gene expression in activated HSCs. Tcf3 regulation of differential gene expression in activated HSCs may be supported by studies investigating the antiadipogenic role of Wnt signaling in the profibrogenic response, as a loss of adipogenic transcriptional regulation has been shown to be important for HSC activation.[2] Canonical Wnt signaling normally leads to beta-catenin nuclear translocation and TCF/LEF-dependent transcription of genes such as cyclin D and c-jun. Blocking this pathway results in suppression of proliferation and collagen type I expression, suggesting Wnt signaling, through downstream TCF, as a mediator of HSC activation.[3]
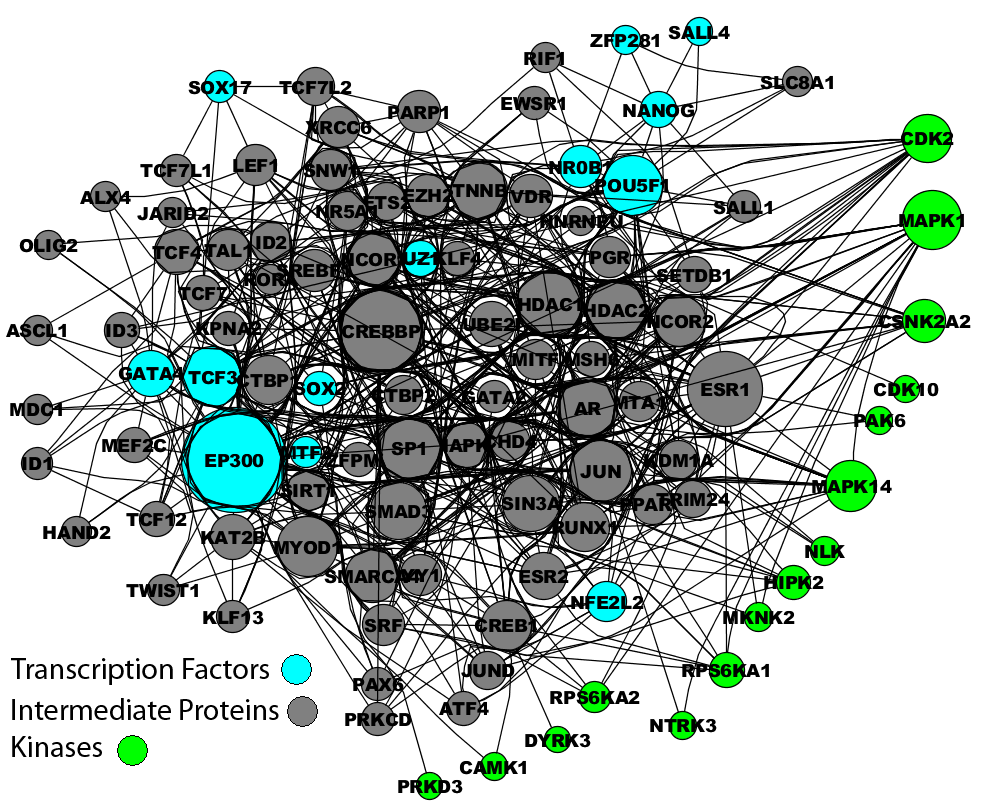
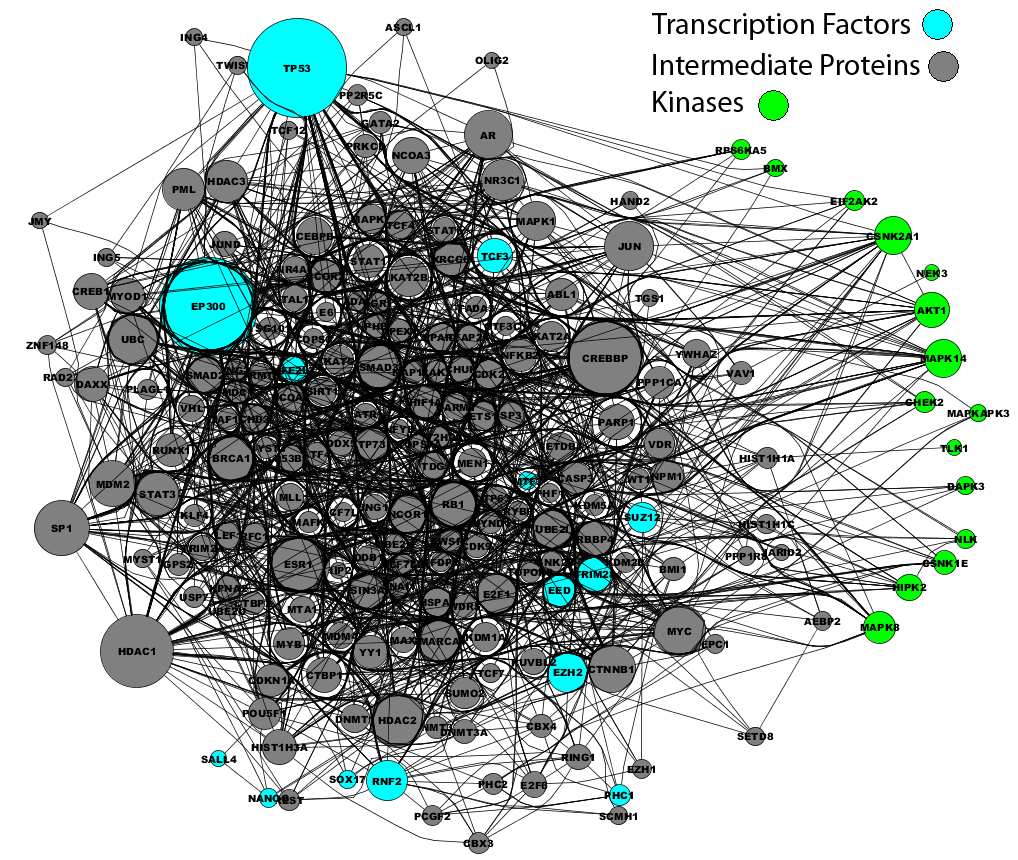
Using the top 15 predicted transcription factors as seeds for building a larger protein complex with X2K, we identified a subnetwork of 95 intermediate proteins that connect the transcription factors using known protein-protein interactions, as well as the most likely protein kinases that phosphorylate the predicted protein complex. Among the predicted kinases is Akt, which is known to be activated downstream of the profibrogenic cannabinoid receptor CB1R to promote HSC survival during liver injury.[4] Akt has also been shown to mediate the anti-apoptotic effect and profibrogenic properties of leptin in HSCs[5], to mediate PDGF-induced proliferation signals, and to regulate collagen type I expression in activated HSCs.[6] Another predicted kinase, RPS6KA6 (RSK4), is a member of the RSK family of serine/threonine kinases that can phosphorylate C/EBPbeta, an adipogenic transcription factor known to regulate collagen type I expression. Rsk-mediated phosphorylation of C/EBPbeta at Thr217 appears to be crucial for the progression of fibrosis; Rsk inhibition led to regression of fibrosis in CCl4-treated mice, and increased activation of RSK and phosphorylated C/EBPbeta both were found in activated HSCs of liver fibrosis patients.[7] Although this study and others examining RSK in HSCs were not isoform-specific, it would be interesting to investigate whether there is distinct contribution of the RSKs we identified to HSC activation.
Zhang et al. [8] examined gene expression changes in neonatal mouse hippocampal neurons undergoing induction of rhythmic network activity. Reanalysis of this data using X2K has recaptured many of the important transcription factors and signaling pathways affected by increases in neural network activity and NMDA receptor signaling. The up-regulated and down-regulated genes from supplemental table S1 from the study were extracted. In young neurons from the hippocampus, NMDA receptor activation, can either promote cell survival, or cell death, depending on the age of the neurons, and whether synaptic or extrasynaptic neurons are activated.[9] The phosphorylation of CREB at a specific residue is crucial for this switch. Activation of a CREB shutoff pathway, which seems to involve CREB Binding Protein (CBP), EP300, and Protein Phosphatase 1 (PP-1) are heavily implicated in the literature.[10] Phosphorylation of CREB in learning and memory contexts has also been described, specifically implicated are Mitogen and stress activated kinase Mnk1, better known as ribosomal S6 kinase.[11] Reanalysis of gene expression modulation caused by increased neural network activity using X2K identified these major regulators discriminatory.
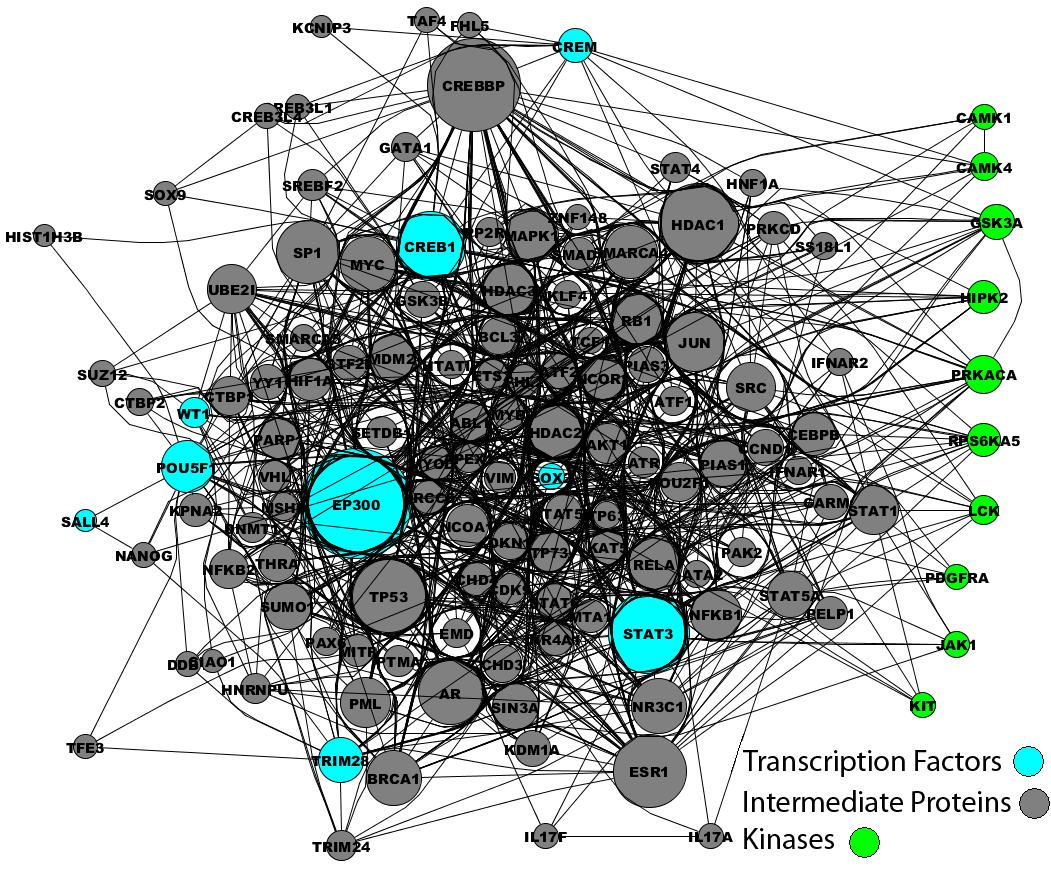
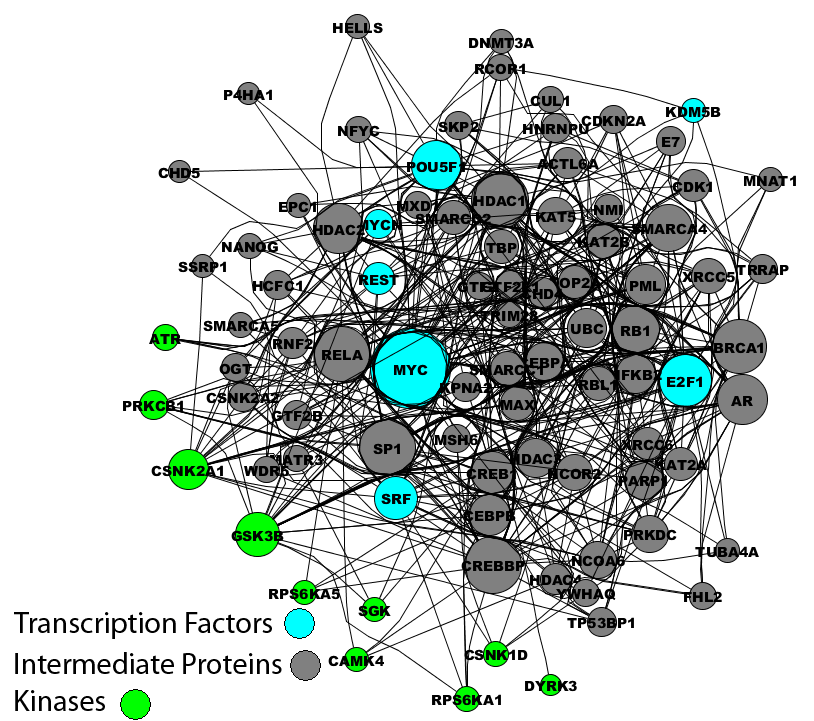
In addition, two calmodulin dependent kinases where identified by X2K applied to the smaller set of genes that were NMDA blockade sensitive: Calcium/calmodulin-dependent protein kinase IV and I (CamK4 and CamK1). The link between CamK4 and CREB dependent transcription is well established.[12,13] Following this link to NMDA receptor activation is clearly through calcium signaling. It was shown that CamK4 activation is important for different forms of LTP that depend on NMDA receptor activation.[14] Activity dependent increases in intracellular calcium, likely through voltage gated calcium channels, affect increases in nuclear calcium where CamK4 is preferentially localized. Hence, the X2K pipeline is capable of recovering known pathways and likely predicting pathways not known to be involved before.
[2]She H, Xiong S, Hazra S, Tsukamoto H: Adipogenic Transcriptional Regulation of Hepatic Stellate Cells. Journal of Biological Chemistry 2005, 280(6):4959-4967.
[3]Cheng JH, She H, Han Y-P, Wang J, Xiong S, Asahina K, Tsukamoto H: Wnt antagonism inhibits hepatic stellate cell activation and liver fibrosis. American Journal of Physiology - Gastrointestinal and Liver Physiology 2008, 294(1):G39-G49.
[4]Teixeira-Clerc F, Julien B, Grenard P, Van Nhieu JT, Deveaux V, Li L, Serriere-Lanneau V, Ledent C, Mallat A, Lotersztajn S: CB1 cannabinoid receptor antagonism: a new strategy for the treatment of liver fibrosis. Nat Med 2006, 12(6):671-676.
[5]Saxena NK, Titus MA, Ding X, Floyd J, Srinivasan S, Sitaraman SV, Anania FA: Leptin as a novel profibrogenic cytokine in hepatic stellate cells: mitogenesis and inhibition of apoptosis mediated by extracellular regulated kinase (Erk) and Akt phosphorylation. The FASEB Journal 2004, 18(13):1612-1614.
[6]Reif S, Lang A, Lindquist JN, Yata Y, Gabele E, Scanga A, Brenner DA, Rippe RA: The Role of Focal Adhesion Kinase-Phosphatidylinositol 3-Kinase-Akt Signaling in Hepatic Stellate Cell Proliferation and Type I Collagen Expression. Journal of Biological Chemistry 2003, 278(10):8083-8090.
[7]Buck M, Chojkier M: A Ribosomal S-6 Kinase Mediated Signal to C/EBP Is Critical for the Development of Liver Fibrosis. PLoS ONE 2007, 2(12):e1372.
[8]Zhang S-J, Steijaert MN, Lau D, Sch端tz G, Delucinge-Vivier C, Descombes P, Bading H: Decoding NMDA Receptor Signaling: Identification of Genomic Programs Specifying Neuronal Survival and Death. Neuron 2007, 53(4):549-562.
[9]Hardingham GE, Bading H: Coupling of extrasynaptic NMDA receptors to a CREB shut-off pathway is developmentally regulated. Biochimica et Biophysica Acta (BBA) - Proteins & Proteomics 2002, 1600(1-2):148-153.
[10]Hardingham GE, Chawla S, Cruzalegui FH, Bading H: Control of Recruitment and Transcription-Activating Function of CBP Determines Gene Regulation by NMDA Receptors and L-Type Calcium Channels. Neuron 1999, 22(4):789-798.
[11]Banko JL, Hou L, Klann E: NMDA receptor activation results in PKA- and ERK-dependent Mnk1 activation and increased eIF4E phosphorylation in hippocampal area CA1. Journal of Neurochemistry 2004, 91(2):462-470.
[12]Matthews RP, Guthrie CR, Wailes LM, Zhao X, Means AR, McKnight GS: Calcium/calmodulin-dependent protein kinase types II and IV differentially regulate CREB-dependent gene expression. Mol Cell Biol 1994, 14(9):6107-6116.
[13]Sun P, Enslen H, Myung PS, Maurer RA: Differential activation of CREB by Ca2+/calmodulin-dependent protein kinases type II and type IV involves phosphorylation of a site that negatively regulates activity. Genes & Development 1994, 8(21):2527-2539.
[14]Kang H, Sun LD, Atkins CM, Soderling TR, Wilson MA, Tonegawa S: An Important Role of Neural Activity-Dependent CaMKIV Signaling in the Consolidation of Long-Term Memory. Cell 2001, 106(6):771-783.